A class of variance estimation methods uses replication. These methods repeatedly estimate a statistic using a slightly different sample. Each sample is called a "replicate." These methods estimate the variance of a target statistic as a function of the empirical variation among estimates from the replicates.
If you are interested in the details of a specific statistical model, rather than the variance estimation method, you can see the procedure directly. Currently AM offers replication variance estimation for the following procedures:
In many data sets, replicates are embodied in a set of replicate weights. These weights adjust the effective sample to correspond to the appropriate replicates. Variance estimates may be obtained by repeatedly reestimating the statistic, each time using a different replicate weight.
There are three general replication procedures that are in wide use:
Jackknife variance estimation typically proceeds by dropping one primary sampling unit for each replicate. In complex designs, the sampling weights associated with the dropped units are then redistributed in a fashion that makes sense given the design. For example, in a stratified design, the weights would then be redistributed within the strata.
Balanced repeated replication divides the set of primary sampling units into an exhaustive set of half-sample replicates. Again, the exact procedures depend on the sample design.
Bootstrap replication resamples from the sample with replacement mimicking the original sample design.
For details of the statistical procedures, see the procedures themselves:
For details of the variance estimation procedures, see them:
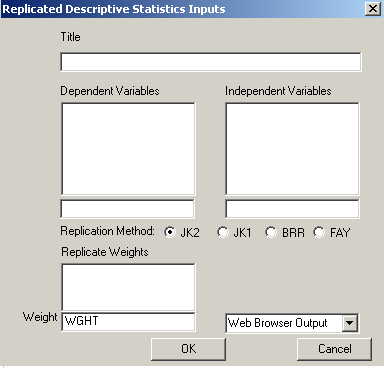
This is a sample replication dialog box--this one for the replicated descriptive statistics. Dependent and independent variables are specified in the usual manner. If an overall weight is used, it goes in the edit box labeled "weight." Replicate weights are dragged into the "replicate weights" box. If default replicate weights are specified, they will appear in the box automatically.
The user must specify the replication method. It is important that you correctly identify the replication method for which the weights were developed, or the estimates will not be correct.
Jackknife 2 (JK2) may be the most common method, which is employed when the sample was explicitly stratified. JK1 uses a slightly different reweighting factor and is typically used when the sample was not explicitly stratified. Estimates for these two methods tend to be very similar, so it will not be immediately apparent if the wrong method is specified. Balanced Repeated Replication estimates are much smaller than the jackknife estimates, so you've used BRR and the standard error numbers are unreasonably small your weights were probably developed for a jackknife procedure. Similarly, if you've selected a jackknife procedure and your standard errors are unreasonably large, the weights were probably developed for BRR. Fay's method is a variant of the BRR.
If you select Fay's method (which you should only do if your weights were developed for Fay's method) you will have to supply an additional piece of information--the re-weighting factor (a number between 0 and 1 that represents the adjustment to the weights of each half sample).
When you select Fay's method, an edit box will appear that will allow you to enter the constant:
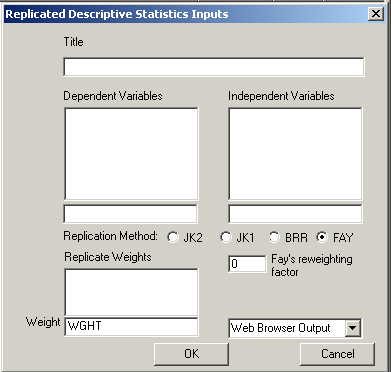
Typically NAEP has used jackknife variance estimation. Specifically, the JK2 variant.